PARP1pred: a web server for screening the bioactivity of inhibitors against DNA repair enzyme PARP-1
DOI:
https://doi.org/10.17179/excli2022-5602Keywords:
PARP-1, DNA repair, machine learning, QSAR, webserver, cheminformaticsAbstract
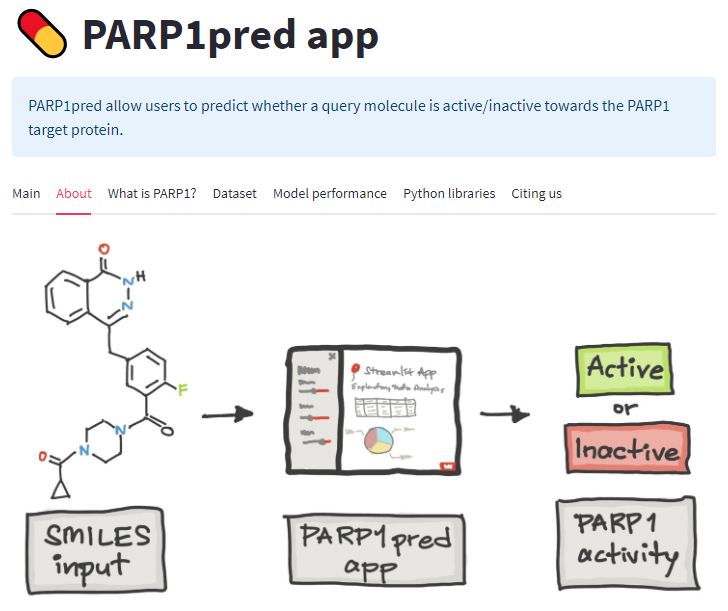
Cancer is the leading cause of death worldwide, resulting in the mortality of more than 10 million people in 2020, according to Global Cancer Statistics 2020. A potential cancer therapy involves targeting the DNA repair process by inhibiting PARP-1. In this study, classification models were constructed using a non-redundant set of 2018 PARP-1 inhibitors. Briefly, compounds were described by 12 fingerprint types and built using the random forest algorithm concomitant with various sampling approaches. Results indicated that PubChem with an oversampling approach yielded the best performance, with a Matthews correlation coefficient > 0.7 while also affording interpretable molecular features. Moreover, feature importance, as determined from the Gini index, revealed that the aromatic/cyclic/heterocyclic moiety, nitrogen-containing fingerprints, and the ether/aldehyde/alcohol moiety were important for PARP-1 inhibition. Finally, our predictive model was deployed as a web application called PARP1pred and is publicly available at https://parp1pred.streamlitapp.com, allowing users to predict the biological activity of query compounds using their SMILES notation as the input. It is anticipated that the model described herein will aid in the discovery of effective PARP-1 inhibitors.
Downloads
Published
How to Cite
License
Copyright (c) 2023 Tassanee Lerksuthirat, Sermsiri Chitphuk, Wasana Stitchantrakul, Donniphat Dejsuphong, Aijaz Ahmad Malik, Chanin Nantasenamat

This work is licensed under a Creative Commons Attribution 4.0 International License.
Authors who publish in this journal agree to the following terms:
- The authors keep the copyright and grant the journal the right of first publication under the terms of the Creative Commons Attribution license, CC BY 4.0. This licencse permits unrestricted use, distribution and reproduction in any medium, provided that the original work is properly cited.
- The use of general descriptive names, trade names, trademarks, and so forth in this publication, even if not specifically identified, does not imply that these names are not protected by the relevant laws and regulations.
- Because the advice and information in this journal are believed to be true and accurate at the time of publication, neither the authors, the editors, nor the publisher accept any legal responsibility for any errors or omissions presented in the publication. The publisher makes no guarantee, express or implied, with respect to the material contained herein.
- The authors can enter into additional contracts for the non-exclusive distribution of the journal's published version by citing the initial publication in this journal (e.g. publishing in an institutional repository or in a book).





