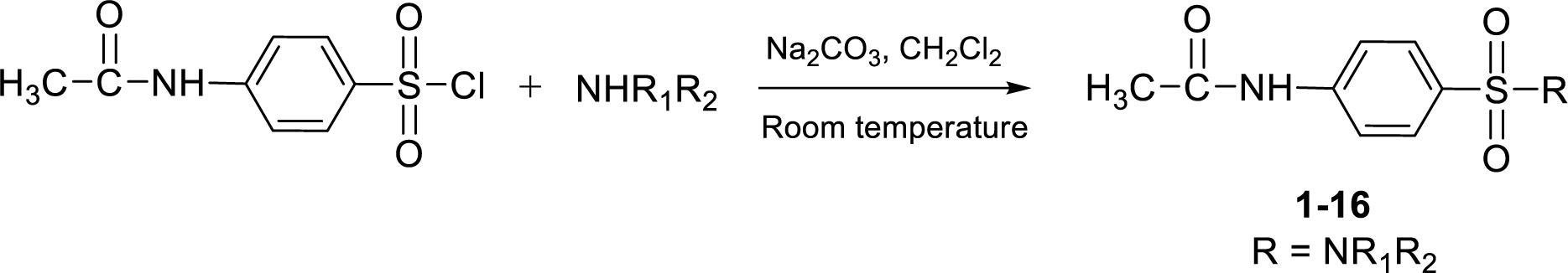Research article
Synthesis of acetamidosulfonamide derivatives with antioxidative and QSAR studies
Apilak Worachartcheewan1[*], Somchai Pisutjaroenpong2, Ratchanok Pingaew3, Supaluk Prachayasittikul4, Suphakit Siriwong1, Somsak Ruchirawat2,5,6, Virapong Prachayasittikul7
1Department of Community Medical Technology, Faculty of Medical Technology, Mahidol University, Bangkok 10700, Thailand2Laboratory of Medicinal Chemistry, Chulabhorn Research Institute, Bangkok 10210, Thailand
3Department of Chemistry, Faculty of Science, Srinakharinwirot University, Bangkok 10110, Thailand
4Center of Data Mining and Biomedical Informatics, Faculty of Medical Technology, Mahidol University, Bangkok 10700, Thailand
5Program in Chemical Science, Chulabhorn Graduate Institute, Bangkok 10210, Thailand
6Center of Excellence on Environmental Health and Toxicology, Commission on Higher Education (CHE), Ministry of Education, Thailand
7Department of Clinical Microbiology and Applied Technology, Faculty of Medical Technology, Mahidol University, Bangkok 10700, Thailand
EXCLI J 2022;21:Doc360
Abstract
A series of sixteen acetamidosulfonamide derivatives (1-16) have been synthesized and investigated for their antioxidant (radical scavenging and superoxide dismutase (SOD)) and antimicrobial activities. Most compounds exhibited antioxidant activities in which compound 15 displayed the most potent radical scavenging and SOD activities. Quantitative structure-activity relationship (QSAR) has been studied using multiple linear regression. The constructed QSAR models displayed high correlation coefficient (Q2LOO-CV = 0.9708 and 0.8753 for RSA and SOD activities, respectively), but low root mean square error (RMSELOO-CV = 0.5105 and 1.3571 for RSA and SOD activities, respectively). The structure-activity relationship showed that an ethylene group connected to pyridine ring provided significant antioxidant activities. The QSAR models give insight into the rational designed of eighty new sulfonamides with various electron donating and withdrawing groups. The top five new designed sulfonamides with nitro group are potential antioxidants to be further developed for medicinal applications.
Keywords: sulfonamides, antioxidants, superoxide dismutase, QSAR, rational design
Introduction
Free radical, a molecule having an unpaired electron, plays physiological signalings in the body such as cell growth, hormone synthesis and killing microorganisms in normal condition (Droge 2002[11]; Knight 2000[18]; Lobo et al., 2010[20]). However, excessive or uncontrolled free radical productions known as oxidative stress cause biological alterations and eventually chronic diseases such as diabetes mellitus, aging, cardiovascular disease, cancer and other degenerative diseases (Pham-Huy et al., 2008[28]; Phaniendra et al., 2015[29]). Therefore, compounds or substances acting as antioxidants have drawn considerable attention in finding new medicinal agents (Carocho and Ferreira, 2013[5]; Wojcik et al., 2010[44]). The target compounds could be achieved based on the known pharmacophores and functional groups of drugs/bioactive compounds (Hughes et al., 2011[15]; Lounnas et al., 2013[21]). A variety of bioactive compounds has been reported i.e., sulfonamide and its derivatives (Khan et al., 2018[17]).
Sulfonamide (SO2NH) is an important functional pharmacophore found in drugs and bioactive compounds. For example, sulfa drugs were used for the prevention and cure of bacterial infections (Smith and Powell, 2000[37]). Chemically modified core structures of sulfonamide have been reported (Bhat et al., 2005[4]; Carta et al., 2012[6]) to improve pharmacological properties such as antimicrobial (Durgun et al., 2017[12]), anticancer (Scozzafava et al., 2003[35]), antiviral (Supuran et al., 2004[38]), and antimalarial (Ugwu et al., 2017[42]) activities as well as cyclooxygenase-2 inhibitors (Gedawy et al., 2020[14]). In addition, 4-substituted (R = NO2, OCH3, CH3, Cl) benzenesulfonamides and related compounds were synthesized and investigated for antioxidant, antimicrobial, anticancer and antifertility activities (Doungsoongnuen et al., 2011[10]; Pholpramool et al., 1991[30]; Temcharoen et al., 1994[40]). Furthermore, sulfonamides containing coumarin moieties were reported to exhibit antioxidant activity (Saeedi et al., 2014[34]).
Based on the literature, 4-acetamidobenzenesulfonamide analogs are attractive target molecules to be explored as antimicrobials, antioxidants and computational study approaching their physicochemical properties related to biological activities.
Quantitative structure-activity relationship (QSAR) has been a common computational technique for model construction using for elucidation of the structure-activity relationship (SAR) of compounds (Nantasenamat et al., 2010[22]; Prachayasittikul et al., 2015[33]).
This study aims to synthesize acetamidosulfonamide derivatives and investigate for antioxidant and antimicrobial activities as well as QSAR study using multiple linear regression (MLR).
Materials and Methods
Chemistry
Column chromatography was carried out using silica gel 60 (70-230 mesh ASTM). Analytical thin-layer chromatography (TLC) was performed on silica gel 60 F254 aluminum sheets. 1H- and 13C-NMR spectra were recorded on a Bruker AVANCE 300 NMR spectrometer. FTIR spectra were obtained using a universal attenuated total reflectance attached on a Perkin-Elmer Spectrum One spectrometer. High resolution mass spectra (HRMS) were recorded on a Bruker Daltonics (microTOF). Melting points were determined using a Griffin melting point apparatus and were uncorrected. Chemicals and reagents were purchased: vitamin E, DPPH (2,2-diphenyl-1-picrylhydrazyl), nitro blue tetrazolium (NBT) salt, L-methionine, riboflavin, Triton-100 and superoxide dismutase (SOD, bovine erythrocytes) from Sigma, USA. DMSO (dimethyl sulfoxide, 99.9 %) from RCI Labscan, Thailand; methanol from Merck, Germany; ampicillin, ciprofloxacin and tetracycline from Sigma, USA; Muller Hinton Broth and Muller Hinton Agar from Becton Dickinson, USA; and sodium chloride from Merck, German. Solvents were analytical grades.
General procedure for the synthesis of sulfonamides (1-16)
A solution of 4-acetamidobenzenesulfonyl chloride (5 mmol) in dichloromethane (30 mL) was added in a dropwise manner to a stirred mixture of amine (5 mmol) and sodium carbonate (7 mmol) in dichloromethane (20 mL). The reaction mixture was stirred at room temperature until completion of reaction (monitored by TLC), and added distilled water (20 mL). The organic phase was separated and the aqueous phase was extracted with dichloromethane (2 × 30 mL). The organic extracts were combined and washed with water (30 mL). The organic layer was dried over anhydrous sodium sulfate (anh. Na2SO4), filtered and evaporated to dryness under reduced pressure. The crude product was further purified by column chromatography on silica gel. In case of sulfonamide 7, it was synthesized using 4-acetamidobenzenesulfonyl chloride (10 mmol), piperazine (5 mmol) and sodium carbonate (14 mmol).
N-(4-(N-benzylsulfamoyl)phenyl)acetamide (1) (De Luca and Giacomelli, 2008)
White solid. 85 % yield; mp 150-152 oC; IR (UATR) cm-1: 3329, 3271, 1681, 1592, 1531, 1320, 1154. 1H NMR (300 MHz, DMSO-d6) δ 2.08 (s, 3H, CH3CO), 3.93 (d, J = 6.2 Hz, 2H, CH2N), 7.14-7.31 (m, 5H, ArH), 7.71 (d, J = 9.4 Hz, 2H, ArH), 7.73 (d, J = 9.4 Hz, 2H, ArH), 7.99 (t, J = 6.2 Hz, 1H, NHSO2), 10.30 (s, 1H, NHCO). 13C NMR (75 MHz, DMSO-d6) δ 23.9, 46.0, 118.5, 126.9, 127.4, 128.0, 134.4, 137.6, 142.5, 168.8. TOF-MS m/z: 305.0968 (Calcd for C15H17N2O3S: 305.0954).
N-(4-(N-(pyridin-2-ylmethyl)sulfamoyl)phenyl)acetamide (2)
Pale brown solid. 70 % yield; mp 116-118 °C; IR (UATR) cm-1: 3330, 3232, 1673, 1592, 1534, 1319, 1151. 1H NMR (300 MHz, DMSO-d6) δ 1.99 (s, 3H, CH3CO), 3.96 (d, J = 6.3 Hz, 2H, CH2N), 7.14 (dd, J = 7.4, 5.0 Hz, 1H, PyH), 7.26 (d, J = 7.8 Hz, 1H, PyH), 7.58-7.70 (m, 5H, ArH4, PyH), 8.03 (t, J = 6.3 Hz, 1H, NHSO2), 8.34 (d, J = 4.7 Hz, 1H, PyH), 10.21 (s, 1H, NHCO). 13C NMR (75 MHz, DMSO-d6) δ 24.6, 48.4, 119.0, 122.1, 122.8, 128.1, 134.6, 137.1, 143.2, 149.2, 157.7, 169.4. TOF-MS m/z: 328.0731 (Calcd for C14H15N3NaO3S: 328.0726).
N-(4-(N-(pyridin-3-ylmethyl)sulfamoyl)phenyl)acetamide (3)
Pale yellow solid. 74 % yield; mp 179-180 oC; IR (UATR) cm-1: 3178, 1681, 1591, 1543, 1322, 1159. 1H NMR (300 MHz, DMSO-d6) δ 2.08 (s, 3H, CH3CO), 3.99 (s, 2H, CH2N), 7.28 (dd, J = 7.7, 4.8 Hz, 1H, PyH), 7.62 (d, J = 7.8 Hz, 1H, PyH), 7.70 (d, J = 9.3 Hz, 2H, ArH), 7.73 (d, J = 9.4 Hz, 2H, ArH), 8.36-8.45 (m, 2H, PyH), 10.33 (br s, 1H, NHCO). 13C NMR (75 MHz, DMSO-d6) δ 24.1, 43.7, 118.6, 123.3, 127.7, 133.4, 134.2, 135.4, 142.8, 148.3, 148.9, 169.0. TOF-MS m/z: 328.0724 (Calcd for C14H15N3NaO3S: 328.0726).
N-(4-(N-((tetrahydrofuran-2-yl)methyl)sulfamoyl)phenyl)acetamide (4)
White solid. 79 % yield; mp 130-132 oC; IR (UATR) cm-1: 3329, 1691, 1592, 1533, 1315, 1152. 1H NMR (300 MHz, DMSO-d6) δ 1.40-1.88 (m, 4H, 2 × CH2), 2.06 (s, 3H, CH3CO), 2.72 (t, J = 6.2 Hz, 2H, CH2N), 3.47-3.81 (m, 3H, CH2O, CHO), 7.56 (t, J = 6.2 Hz, 1H, NHSO2), 7.69 (d, J = 9.0 Hz, 2H, ArH), 7.73 (d, J = 9.2 Hz, 2H, ArH), 10.31 (s, 1H, NHCO). 13C NMR (75 MHz, DMSO-d6) δ 24.1, 25.1, 28.5, 46.6, 67.3, 77.0, 118.7, 127.7, 134.5, 142.7, 169.1. TOF-MS m/z: 299.1055 (Calcd for C13H19N2O4S: 299.1060).
N-(4-((4-methylpiperazin-1-yl)sulfonyl)- phenyl)acetamide (5) (Barbosa et al., 2009)
White solid. 78 % yield; mp 286-287 oC; IR (UATR) cm-1: 3327, 1682, 1590, 1530, 1316, 1151. 1H NMR (300 MHz, DMSO-d6) δ 2.07 (s, 3H, CH3CO), 2.10 (s, 3H, CH3N), 2.32 (t, J = 4.7 Hz, 4H, 2 × CH2), 2.83 (br t, 4H, 2 × CH2), 7.63 (d, J = 8.9 Hz, 2H, ArH), 7.80 (d, J = 8.8 Hz, 2H, ArH), 10.43 (s, 1H, NHCO). 13C NMR (75 MHz, DMSO-d6) δ 24.2, 45.4, 45.8, 53.6, 118.8, 128.8, 128.9, 143.6, 169.4. TOF-MS m/z: 298.1225 (Calcd for C13H20N3O3S: 298.1220).
N-(4-((4-benzylpiperazin-1-yl)sulfonyl)phenyl)acetamide (6)
White solid. 81 % yield; mp 189-190 oC; IR (UATR) cm-1: 3337, 1680, 1590, 1528, 1314, 1160. 1H NMR (300 MHz, DMSO-d6) δ 2.18 (s, 3H, CH3CO), 2.49 (br t, 4H, 2 × CH2), 2.93 (br t, 4H, 2 × CH2), 3.53 (s, 2H, CH2Ph), 7.25-7.40 (m, 5H, ArH), 7.73 (d, J = 8.8 Hz, 2H, ArH), 7.90 (d, J = 8.8 Hz, 2H, ArH), 10.48 (s, 1H, NHCO). 13C NMR (75 MHz, DMSO-d6) δ 24.0, 45.8, 51.3, 61.3, 118.6, 126.9, 128.1, 128.6, 128.7, 137.6, 143.4, 169.0. TOF-MS m/z: 374.1531 (Calcd for C19H24N3O3S: 374.1533).
N,N'-((piperazine-1,4-disulfonyl)bis(4,1-phenylene))diacetamide (7)
White solid. 66 % yield; mp 309-310 oC; IR (UATR) cm-1: 3265, 1684, 1588, 1538, 1345, 1160. 1H NMR (300 MHz, DMSO-d6) δ 2.09 (s, 6H, 2 × CH3CO), 2.92 (s, 8H, 4 × CH2), 7.61 (d, J = 8.7 Hz, 4H, ArH), 7.79 (d, J = 8.7 Hz, 4H, ArH), 10.39 (s, 2H, 2 × NHCO). 13C NMR (75 MHz, DMSO-d6) δ 24.2, 45.2, 118.8, 128.3, 128.8, 143.7, 169.3. TOF-MS m/z: 503.1032 (Calcd for C20H24N4NaO6S2: 503.1030).
N-(4-(morpholinosulfonyl)phenyl)- acetamide (8) (Barbosa et al., 2009)
White solid. 73 % yield; mp 159-160 oC; IR (UATR) cm-1: 3303, 1677, 1590, 1526, 1343, 1161. 1H NMR (300 MHz, DMSO-d6) δ 2.08 (s, 3H, CH3CO), 2.81 (t, J = 4.6 Hz, 4H, 2 × CH2), 3.60 (t, J = 4.6 Hz, 4H, 2 × CH2), 7.66 (d, J = 8.8 Hz, 2H, ArH), 7.83 (d, J = 8.8 Hz, 2H, ArH), 10.39 (s, 1H, NHCO). 13C NMR (75 MHz, DMSO-d6) δ 24.2, 45.9, 65.3, 118.7, 127.8, 128.9, 143.6, 169.2. TOF-MS m/z: 307.0734 (Calcd for C12H16N2NaO4S: 307.0723).
N-(4-((3,4-dihydroisoquinolin-2(1H)-yl)sulfonyl)phenyl)acetamide (9) (Pagliero et al., 2011)
White solid. 73 % yield; mp 181-182 oC; IR (UATR) cm-1: 3301, 1678, 1590, 1527, 1337, 1160. 1H NMR (300 MHz, DMSO-d6) δ 2.07 (s, 3H, CH3CO), 2.83 (t, J = 5.6 Hz, 2H, C4-IQH), 3.24 (t, J = 5.8 Hz, 2H, C3- IQH), 4.14 (s, 2H, C1- IQH), 7.05-7.15 (m, 4H, IQH), 7.73 (d, J = 8.9 Hz, 2H, ArH), 7.79 (d, J = 9.0 Hz, 2H, ArH), 10.35 (s, 1H, NHCO). 13C NMR (75 MHz, DMSO-d6) δ 24.6, 28.5, 44.0, 47.7, 119.2, 126.6, 126.9, 127.1, 129.1, 129.8, 132.1, 133.5, 143.9, 169.6. TOF-MS m/z: 353.0942 (Calcd for C17H18N2NaO3S: 353.0942).
N-(4-(N-cyclohexylsulfamoyl)phenyl)- acetamide (10)
White solid. 70 % yield; mp 213-215 oC; IR (UATR) cm-1: 3328, 3273, 1694, 1596, 1539, 1312, 1147. 1H NMR (300 MHz, DMSO-d6) δ 0.90-1.60 (m, 10H, 5 × CH2), 2.07 (s, 3H, CH3CO), 2.86 (br s, 1H, CHN), 7.47 (d, J = 7.3 Hz, 1H, NHSO2), 7.72 (s, 4H, ArH), 10.30 (s, 1H, NHCO). 13C NMR (75 MHz, DMSO-d6) δ 24.1, 24.4, 24.9, 33.3, 52.1, 118.7, 127.4, 136.0, 142.5, 169.1. TOF-MS m/z: 319.1096 (Calcd for C14H20N2NaO3S: 319.1087).
N-(4-(N-cyclopentylsulfamoyl)phenyl)- acetamide (11)
White solid. 69 % yield; mp 179-180 oC; IR (UATR) cm-1: 3274, 1672, 1594, 1539, 1321, 1161. 1H NMR (300 MHz, DMSO-d6) δ 1.15-1.64 (m, 8H, 4 × CH2), 2.07 (s, 3H, CH3CO), 3.25-3.35 (m, 1H, CHN), 7.47 (d, J = 7.1 Hz, 1H, NHSO2), 7.70 (d, J = 9.2 Hz, 2H, ArH), 7.74 (d, J = 9.2 Hz, 2H, ArH), 10.29 (s, 1H, NHCO). 13C NMR (75 MHz, DMSO-d6) δ 23.2, 24.5, 33.1, 54.7, 119.1, 128.1, 135.7, 142.9, 169.7. TOF-MS m/z: 305.0937 (Calcd for C13H18N2NaO3S: 305.0930).
N-(4-(N-phenylsulfamoyl)phenyl)acetamide (12) (Silveira et al., 2017)
White solid. 70 % yield; mp 200-202 oC; IR (UATR) cm-1: 3239, 1672, 1591, 1542, 1333, 1166. 1H NMR (300 MHz, DMSO-d6) δ 2.04 (s, 3H, CH3CO), 6.99 (t, J = 7.1 Hz, 1H, ArH), 7.04 (d, J = 8.3 Hz, 2H, ArH), 7.19 (t, J = 7.7 Hz, 2H, ArH), 10.28 (s, 1H, NHCO). 13C NMR (75 MHz, DMSO-d6) δ 24.4, 119.1, 120.6, 124.6, 128.4, 129.6, 133.5, 138.1, 143.4, 169.8. TOF-MS m/z: 291.0807 (Calcd for C14H15N2O3S: 291.0798).
N-(4-(N-phenethylsulfamoyl)phenyl)- acetamide (13)
White solid. 85 % yield; mp 142-144 oC; IR (UATR) cm-1: 3325, 3273, 1677, 1592, 1530, 1318, 1152. 1H NMR (300 MHz, DMSO-d6) δ 2.04 (s, 3H, CH3CO), 2.60 (t, J = 7.4 Hz, 2H, CH2Ph), 2.88 (q, J = 6.8 Hz, 2H, CH2N), 7.02-7.25 (m, 5H, ArH), 7.49 (d, J = 7.3 Hz, 1H, NHSO2), 7.65 (s, 4H, ArH), 10.36 (s, 1H, NHCO). 13C NMR (75 MHz, DMSO-d6) δ 24.2, 35.4, 44.4, 119.6, 126.8, 128.1, 128.8, 129.0, 134.4, 138.9, 142.7, 170.7. TOF-MS m/z: 341.0927 (Calcd for C16H18N2NaO3S: 341.0930).
N-(4-(N-(3,4-dimethoxyphenethyl)- sulfamoyl)phenyl)acetamide (14)
White solid. 86 % yield; mp 134-136 oC; IR (UATR) cm-1: 3327, 1682, 1592, 1516, 1322, 1149. 1H NMR (300 MHz, DMSO-d6) δ 2.07 (s, 3H, CH3CO), 2.56 (t, J = 7.4 Hz, 2H, CH2Ph), 2.90 (q, J = 6.9 Hz, 2H, CH2N), 3.68, 3.69 (2s, 6H, 2 × OCH3), 6.62 (dd, J = 8.1, 1.6 Hz, 1H, ArH), 6.70 (d, J = 1.6 Hz, 1H, ArH), 6.79 (d, J = 8.2 Hz, 1H, ArH), 7.52 (t, J = 5.7 Hz, 1H, NHSO2), 7.68 (d, J = 9.0 Hz, 2H, ArH), 7.73 (d, J = 9.0 Hz, 2H, ArH), 10.31 (s, 1H, NHCO). 13C NMR (75 MHz, DMSO-d6) δ 24.5, 35.2, 44.7, 55.9, 56.0, 112.4, 113.0, 119.1, 121.0, 128.1, 131.6, 134.6, 143.1, 147.8, 149.1, 169.5. TOF-MS m/z: 401.1145 (Calcd for C18H22N2NaO5S: 401.1142).
N-(4-(N-(2-(pyridin-2-yl)ethyl)sulfamoyl)phenyl)acetamide (15)
White solid. 78 % yield; mp 138-140 oC; IR (UATR) cm-1: 3238, 1686, 1592, 1541, 1319, 1164. 1H NMR (300 MHz, DMSO-d6) δ 2.07 (s, 3H, CH3CO), 2.81 (t, J = 7.4 Hz, 2H, CH2Py), 3.06 (q, J = 6.8 Hz, 2H, CH2N), 7.15-7.22 (m, 2H, PyH), 7.58 (t, J = 5.8 Hz, 1H, NHSO2), 7.62-7.77 (m, 5H, ArH4, PyH), 8.42 (d, J = 4.2 Hz, 1H, PyH), 10.31 (s, 1H, NHCO). 13C NMR (75 MHz, DMSO-d6) δ 24.5, 37.7, 42.7, 119.2, 122.1, 123.7, 128.1, 134.5, 136.9, 143.1, 149.5, 158.8, 169.5. TOF-MS m/z: 320.1055 (Calcd for C15H18N3O3S: 320.1063).
N-(4-(N-(2-(diethylamino)ethyl)- sulfamoyl)phenyl)acetamide (16)
White solid. 71 % yield; mp 76-78 oC; IR (UATR) cm-1: 3322, 3273, 1681, 1592, 1532, 1317, 1153. 1H NMR (300 MHz, DMSO-d6) δ 0.84 (t, J = 7.2 Hz, 6H, 2 × CH3), 2.06 (s, 3H, CH3CO), 2.29-2.40 (m, 6H, 3 × CH2), 2.74 (t, J = 7.3 Hz, 2H, CH2), 7.69 (d, J = 9.0 Hz, 2H, ArH), 7.74 (d, J = 9.1 Hz, 2H, ArH), 10.33 (s, 1H, NHCO). 13C NMR (75 MHz, DMSO-d6) δ 12.2, 24.4, 41.4, 47.1, 52.3, 119.3, 128.0, 135.0, 143.1, 169.5. TOF-MS m/z: 314.1528 (Calcd for C14H24N3O3S: 314.1533).
Biological activity evaluation
Antioxidant activity assays
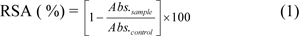
Radical scavenging activity (RSA) was performed by adding 1 mL of 2,2-diphenyl-1-picrylhydrazyl (DPPH) in methanol (0.1 mM) to the tested compounds (dissolved in DMSO) with the final concentration of 300 μg/mL, and the mixture was kept in the dark for 30 min. The absorbance at 517 nm was measured using UV-Visible spectrophotometer (UV-1610, Shimadzu). The DPPH (stable purple compound) reacted with antioxidants to give light-yellow color product of 1,1-diphenyl-2-picrylhydrazine (Worachartcheewan et al., 2012[47]). Vitamin E was used as a control and DMSO was used as a blank reaction. The RSA ( %) was computed using equation (1):
where Abs.control is the absorbance of the control reaction, and Abs.sample is the absorbance of the tested compound.
Superoxide scavenging activity was evaluated using superoxide dismutase (SOD) assay by measuring nitro blue tetrazolium (NBT) reduction (Piacham et al., 2003[31]). The stock solution containing 27 mL of HEPES buffer (50 mM, pH 7.8), 1.5 mL of L-methionine (30 mg/mL), 1 mL of NBT (1.41 mg/mL) and 750 μL of Triton X-100 (1 % wt) was prepared. The 1 mL of stock solution was added to the solution of tested compounds (1-16, dissolved in DMSO) with the final concentration of 300 μg/mL. The reaction was initially started by adding 10 μL of riboflavin (44 mg/mL), and followed by illumination under a Philips Classic Tone lamp (60 W) in a light box for 7 min. The absorbance at 550 nm of the reaction was measured using UV-Visible spectrophotometer (UV-1610, Shimadzu). SOD from bovine erythrocytes was used as a control, and DMSO was used as a blank reaction. The SOD activity (%) was calculated using equation (2):
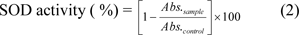
where Abs.control is the absorbance of the control reaction, and Abs.sample is the absorbance of the tested compound.
If compounds exhibited antioxidant (RSA and SOD) activities greater than 50 % at 300 μg/mL, their IC50 values were performed by linear regression plot between %RSA or %SOD activity and compound concentrations.
Antimicrobial activity assay
The tested compounds were investigated for antimicrobial activity using the agar-dilution method (Baron et al., 1994[3]). The compounds were prepared and then transferred to Muller Hinton (MH) agar to obtain the final concentration of 4-256 μg/mL. The microorganisms were cultured in MH broth at 37 °C overnight, and then suspended in 0.9 % normal saline solution. Turbidity of microorganism was adjusted to give a cell density of 1×108 CFU/mL compared to 0.5 McFarland turbidity standard. The microorganisms were inoculated onto the agar plates containing a variety of compound concentrations using a multipoint inoculator, and incubated at 37 °C for 24-48 h. The control plate containing DMSO, MH broth and antibacterial agents (i.e., ampicillin, ciprofloxacin and tetracycline) were also parallelly performed. The inhibition of microbial cell growth was determined as the minimum inhibitory concentration (MIC), which is the lowest concentration to completely inhibit the growth of microorganisms. The twenty-six strains of microorganisms for the assay consisted of reference strains and clinical isolates; gram positive bacteria: Staphylococcus aureus ATCC 29213, Staphylococcus aureus ATCC 25923, Staphylococcus epidermidis ATCC 12228, Enterococcus faecalis ATCC 29212, Enterococcus faecalis ATCC 33186, Micrococcus luteus ATCC 10240, Corynebacterium diphtheriae NCTC 10356, Bacillus subtilis ATCC 6633, Listeria monocytogenes, Bacillus cereus; gram negative bacteria: Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 700603, Serratia marcescens ATCC 8100, Salmonella typhimurium ATCC 13311, Shewanella putrefaciens ATCC 8071, Achromobacter xylosoxidans ATCC 2706, Pseudomonas aeruginosa ATCC 27853, Pseudomonas stutzeri ATCC 17587, Shigella dysenteriae, Salmonella enteritidis, Morganella morganii, Aeromonas hydrophila, Citrobacter freundii, Plesiomonas shigelloides and diploid fungus (yeast): Candida albicans ATCC 90028, Saccharomyces cerevisiae ATCC 2601.
QSAR analysis
Data set
The bioactive sulfonamide derivatives (1-16) were used as a data set to obtain significant descriptors (independent variables) correlating with biological properties (dependent variable) for development of QSAR models.
Calculation of quantum chemical and molecular descriptors
Chemical structures of the sulfonamides were constructed by Chemdraw Pro13 software (PerkinElmer, USA) which subjected to GaussView (Dennington et al., 2003[9]) software and further geometrically optimized by Gaussian 09, Revision A.02 (Frisch et al., 2009[13]) at the semi-empirical level using Austin Model 1 (AM1), then followed by density functional theory (DFT) calculation using Becke's three-parameter hybrid method, and the Lee-Yang-Parr correlation functional (B3LYP) together with the 6-31 g(d) basis set. The low-energy conformers of the compounds were obtained and output files were consequently employed to calculate quantum chemical descriptors including the total energy (Etotal) of the molecule, the highest occupied molecular orbital energy (EHOMO), the lowest unoccupied molecular orbital energy (ELUMO), the total dipole moment (µ) of the molecule, the electron affinity (EA), the ionization potential (IP), the energy difference of HOMO and LUMO (HOMO-LUMOGap), Mulliken electronegativity (χ), hardness (η), softness (S), electrophilicity (ω), electrophilic index (ωi) and the mean absolute atomic charge (Qm) (Karelson et al., 1996[16]; Parr et al., 1978[25], 1999[27]; Parr and Pearson, 1983[26]; Thanikaivelan et al., 2000[41]). The output files were used as the input data for calculating a set of 3,224 molecular descriptors using Dragon software, version 5.5 (Talete, 2007[39]) to give the following 22 categories: 48 constitutional descriptors, 119 topological descriptors, 47 feature selection of descriptors walk and path counts, 33 connectivity indices, 47 information indices, 96 2D autocorrelation, 107 edge adjacency indices, 64 burden eigenvalues, 21 topological charge indices, 44 eigenvalue-based indices, 41 randic molecular profiles, 74 geometrical descriptors, 150 RDF descriptors, 160 3D-MoRSE descriptors, 99 WHIM descriptors, 197 GETAWAY descriptors, 154 functional group counts, 120 atom-centred fragments, 14 charge descriptors, 29 molecular properties, 780 2D binary fingerprints and 780 2D frequency fingerprints.
Feature selection of descriptors
A volume of 3,224 molecular descriptors was filtered to reduce multi-collinear and redundant descriptor variables. Constant values and pairs of variables with correlation coefficient greater than 0.99 were removed in Dragon software. The remaining 1,387 molecular descriptors were combined with a set of 13 quantum chemical descriptors. The descriptors correlated with their activities were selected using automatic variable selection procedure (CfsSubsetEval combined with the BestFirst) in Waikato Environment for Knowledge Analysis (Weka) software, version 3.4.5 (Witten et al., 2011[43]), and following by stepwise multiple linear regression (MLR) (SPSS statistics 18.0, SPSS Inc., USA). The obtained descriptors were determined for the intercorrelation matrix among each descriptor by Pearson's correlation coefficient (r) using SPSS statistics 18.0 (SPSS Inc., USA). The cutoff of |r| ≥ 0.8 was assigned to describe collinearly between the descriptors.
Multiple linear regression for QSAR models construction
QSAR models were carried out by multiple linear regression (MLR) using Weka software, version 3.4.5 (Witten et al., 2011[43]). Significant descriptors were used as independent variables (X) and their biological activities were used as dependent variable (Y). The QSAR models were constructed by equation (3):
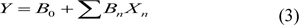
where Y is the biological activities of the compounds, B0 is the intercept, and Bn are the regression coefficients of the descriptors Xn.
Generation of dataset
The data set was divided into two sets composed of training and testing sets. The training set was used to construct the QSAR models, whereas the testing set was employed to evaluate the model. The testing set was performed by means of leave-one-out cross validation (LOO-CV) which one sample was left out from the data set (N) to be used as the testing set while the remaining set (N-1) was used as the training set. This was iteratively continued until every sample had a chance to be the testing set. The biological activities of the compounds were predicted using equation (3).
Evaluation of QSAR models
Predictive performance of the constructed QSAR models was evaluated by statistical analysis. Squared correlation coefficient (R2Tr) for training, and cross-validated Q2 (Q2LOO-CV) for LOO-CV sets were used to measure a relative correlation of the predicted and the experimental values. Furthermore, root mean square error (RMSE) was used to determine the predictive error of the model for training (RMSETr) and LOO-CV (RMSELOO-CV) sets (Nantasenamat et al., 2010[22]).
Results and Discussion
Chemistry
N-sulfonylation
Sixteen sulfonamides (1-16) were synthesized in 66-86 % yields by sulfonylation of various amines with 4-acetamidobenzenesulfonyl chloride containing base, sodium carbonate (Na2CO3), in dichloromethane (CH2Cl2) at room temperature (Figure 1(Fig. 1)).
Structures of the sulfonamides (1-16) were characterized by their HRMS, 1H and 13C NMR and IR spectra. All sulfonamides had molecular ion peaks corresponding to their molecular formula. 1H NMR spectra of N-acetyl compounds (1-16) displayed the characteristic signals of CH3CO and NHCO protons as two singlets at δ in the range of 1.9-2.2 and 10.2-10.5 ppm, respectively whereas their 13C NMR spectra showed carbonyl groups at δ 168-171 ppm. Infrared spectra of the acetamides (1-16) gave N-H and C=O absorptions at around 3170-3340 and 1670-1700 cm−1, respectively.
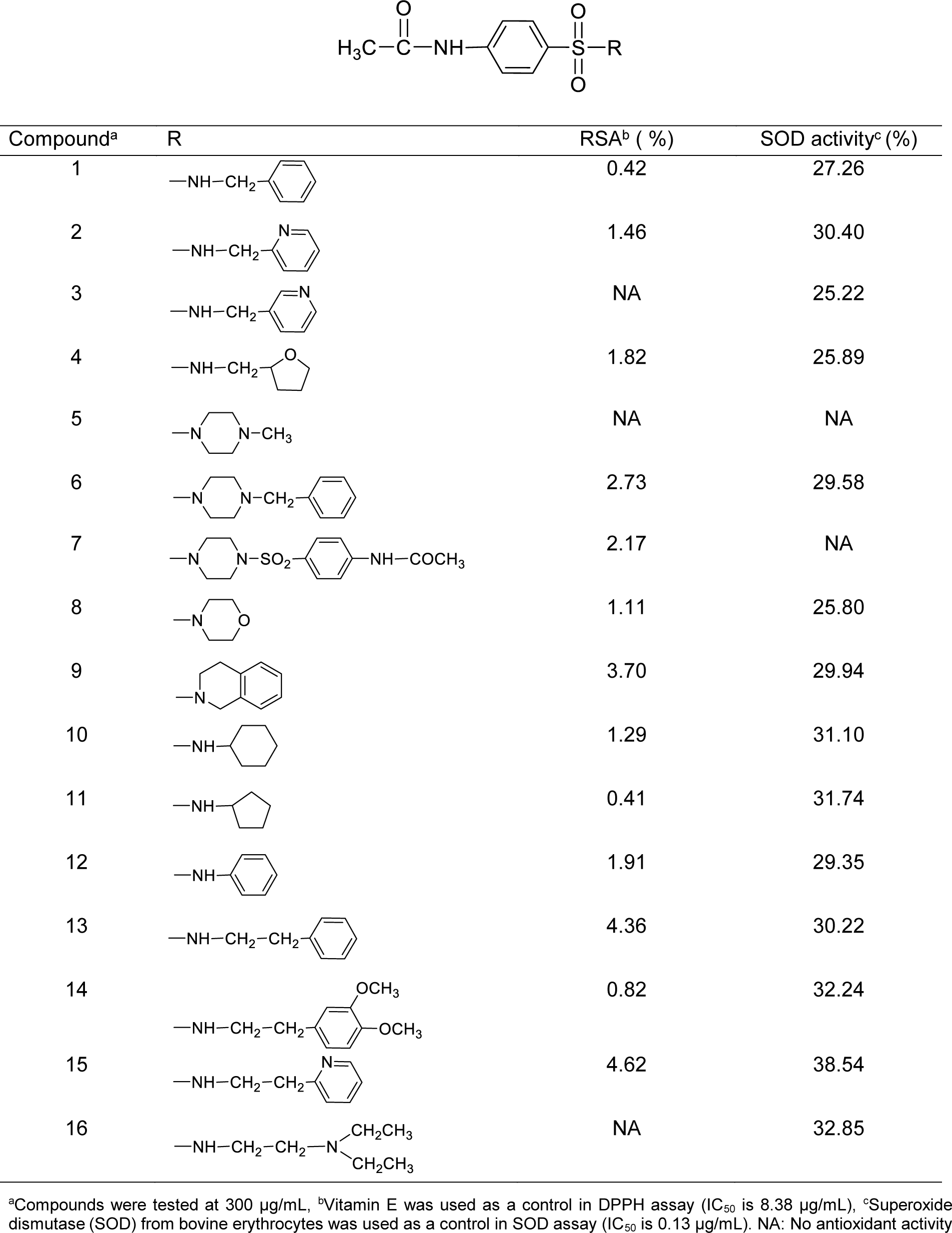
These sulfonamides have a common 4-acetamidobenzenesulfonyl core structure bearing various amino substituents (R) as shown in Table 1(Tab. 1). The R group can be methylamino (1-4, group A), ethylamino (13-16, group B), cyclic amino (5-9, group C) and ring substituted amino (10-12, group D) moieties.
Biological activities
Radical scavenging activity
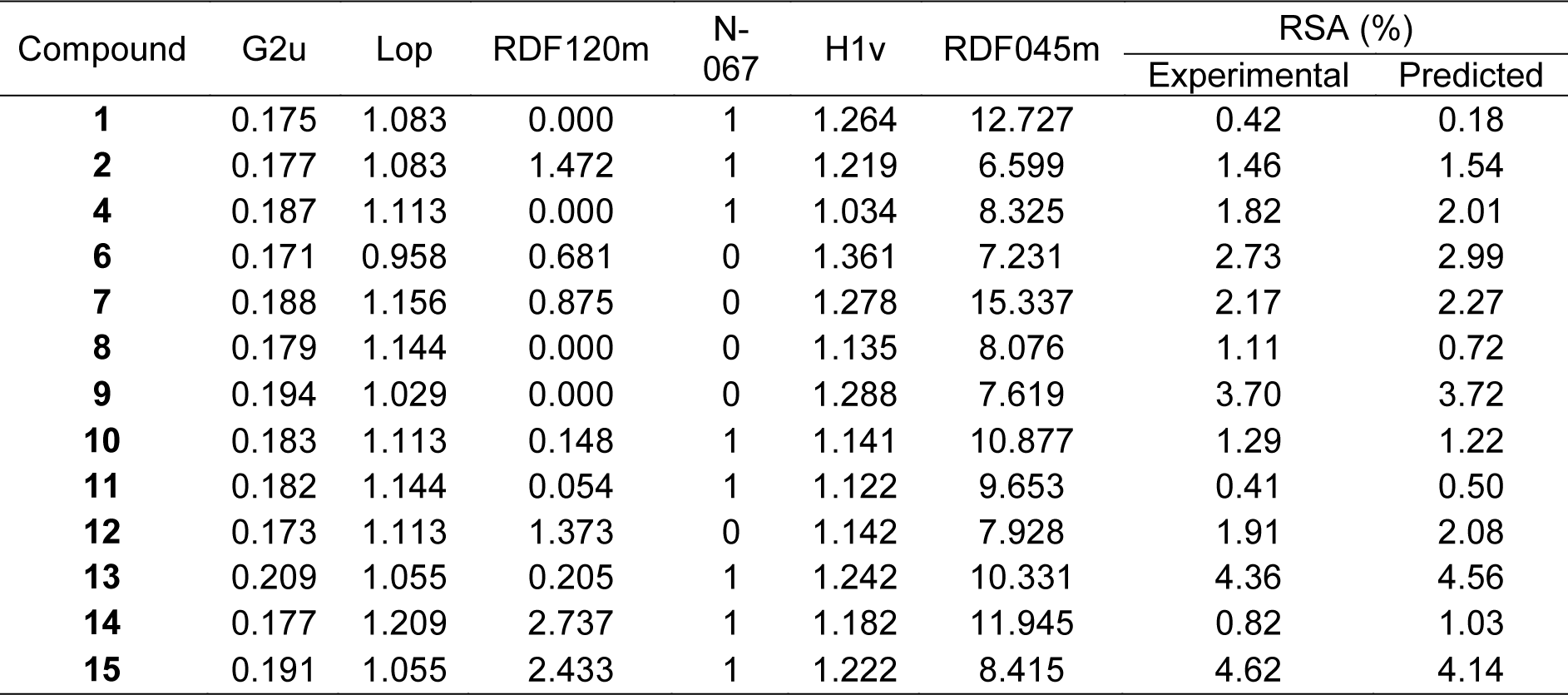
The results revealed that most sulfonamides displayed radical scavenging activity (RSA) toward the DPPH radical in 0.41-4.62 % range at concentration of 300 µg/mL, except for compounds 3, 5 and 16 had no activity (Table 1(Tab. 1)). The activity of compounds is dependent on the functional (R) groups of the core structure (Table 1(Tab. 1)). The most potent compound 15 (R = NHCH2CH2-2-pyridyl) exhibited 4.62 % RSA, whereas R = NHCH2C6H5 (1) showed the lowest activity (0.42 % RSA).
Superoxide scavenging activity
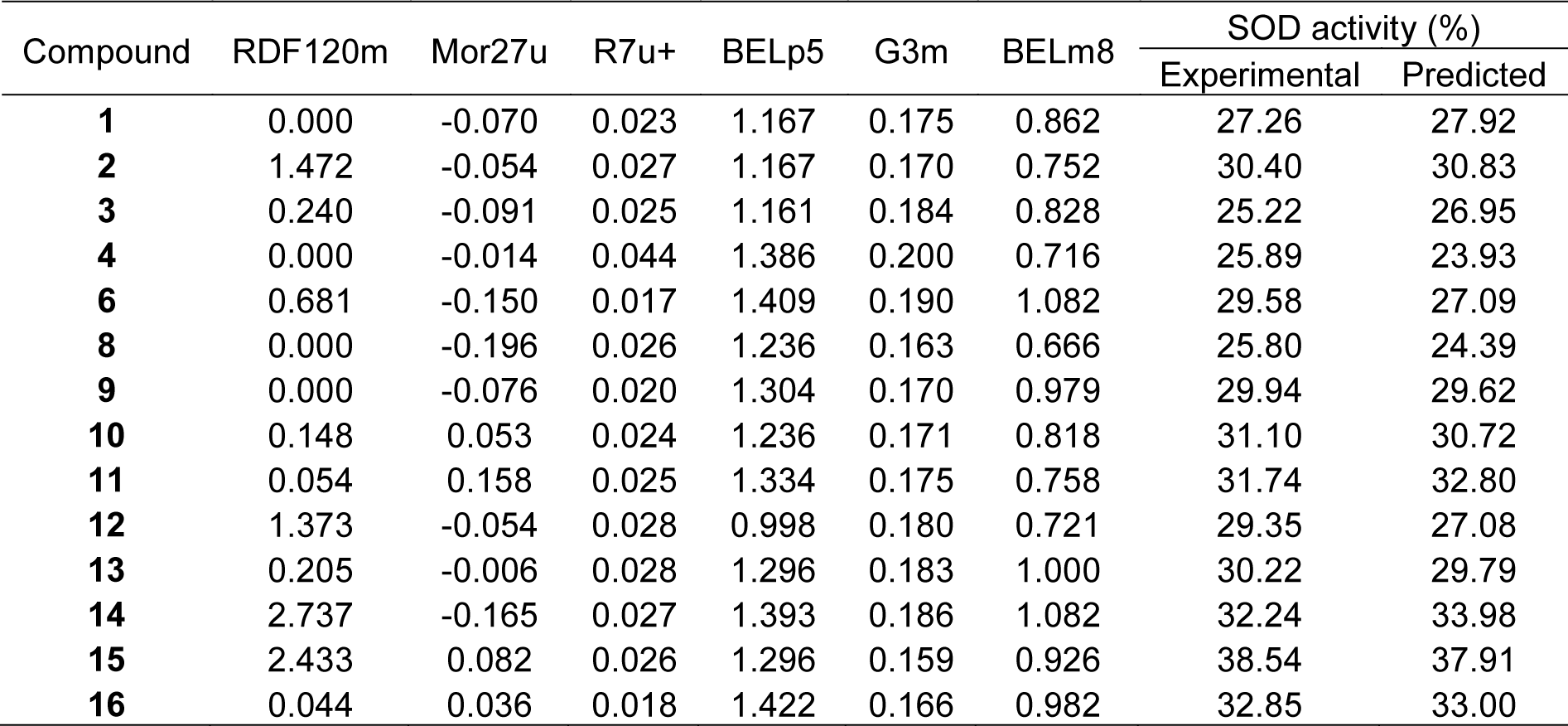
Superoxide scavenging (SOD) assay was performed to show the ability of compounds to inhibit the photoreduction of NBT. Most sulfonamides had the SOD activity (25.22-38.54 %) at 300 µg/mL, except for compounds 5 and 7 with no activity (Table 1(Tab. 1)). Interestingly, compound 15 displayed the highest antioxidant activities in SOD (38.54 %) and DPPH (4.62 %) assays.
Antimicrobial activity
The sulfonamides (1-16) were investigated for antimicrobial activity using the agar dilution against gram positive bacteria, gram negative bacteria and diploid fungi from reference strains and clinical isolates. It was found that all compounds were inactive at concentration of 256 µg/mL.
QSAR analysis
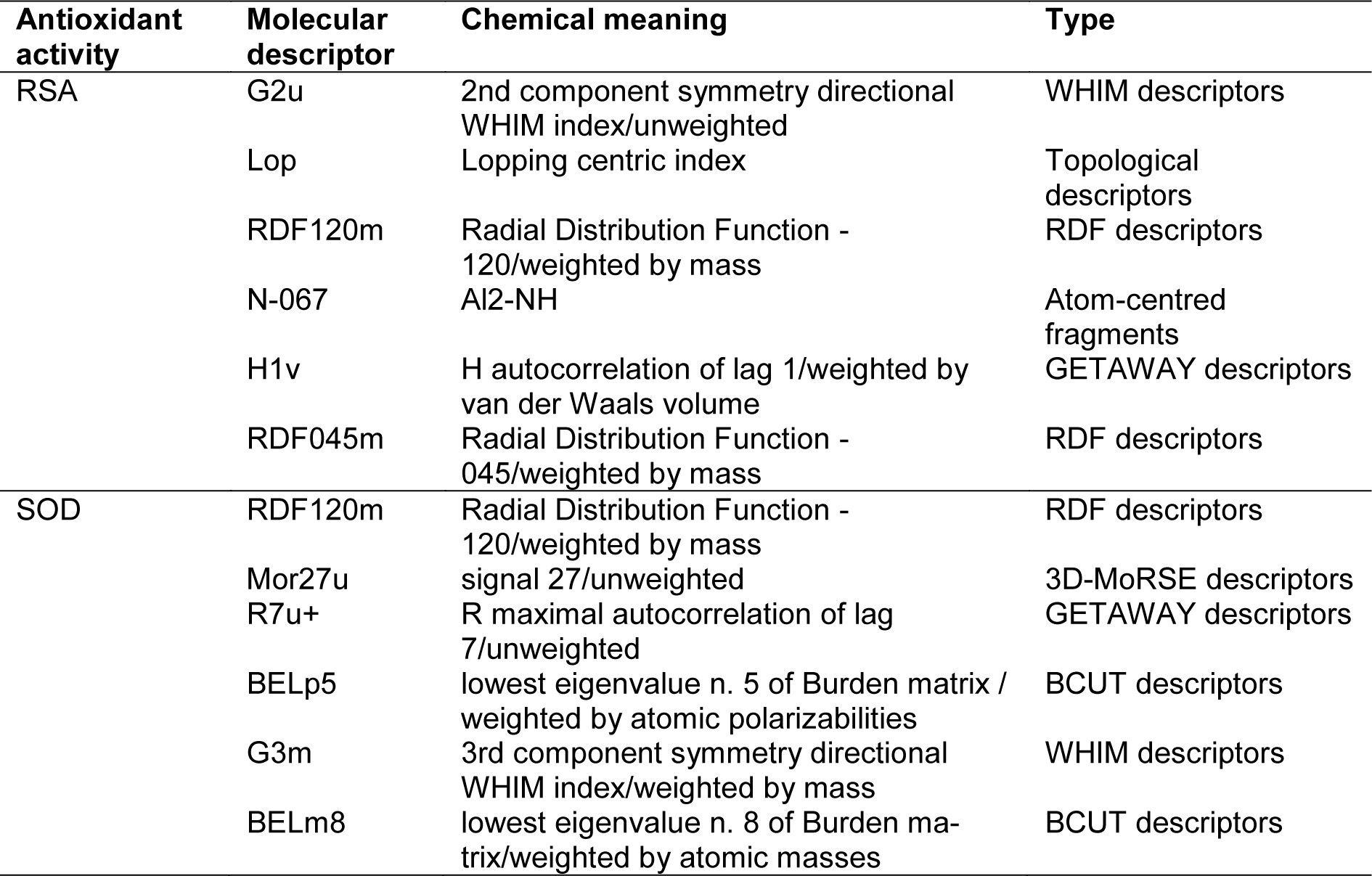
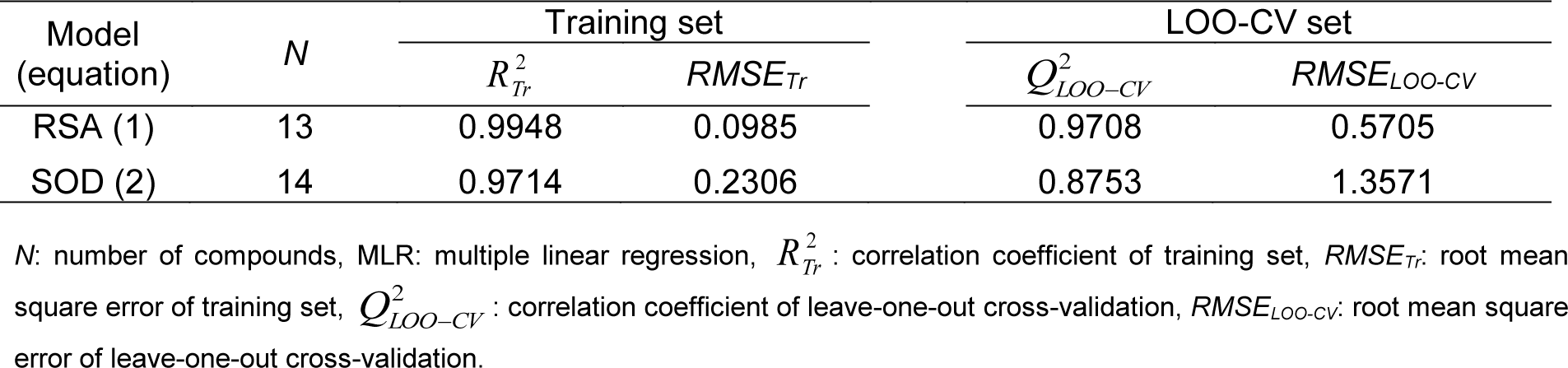
QSAR is a potential model for exploring the relationship between physicochemical properties and bioactivities of compounds as well as rational design of new bioactive compounds. This has been demonstrated in compounds with antimicrobial (Cherdtrakulkiat et al., 2020[7]), antioxidant (Worachartcheewan et al., 2014[46]), anticancer (Leechaisit et al., 2019[19]) and antiviral (Worachartcheewan et al., 2019[48]) activities. To construct the model, the quantum chemical and molecular descriptors of the compounds were computed using Gaussian 09 and Dragon softwares, respectively (Alyar and Karacan, 2009[1]; Nantasenamat et al., 2013[23]; Pingaew et al., 2015[32]; Worachartcheewan et al., 2011[45]). The obtained 3,224 descriptors from the Dragon software were initially filtered to remove multi-collinear and redundant descriptor variables. This process gave 1,387 descriptors which were further combined with 13 quantum chemical descriptors from the Gaussian 09 software to provide the final set of 1,400 descriptors. The important descriptors correlated with their antioxidant activities were achieved using automatic variable selection procedure (CfsSubsetEval combined with the BestFirst) in WEKA software and following by stepwise MLR. Six descriptors for the QSAR model of RSA included G2u, Lop, RDF120m, N-067, H1v and RDF045m, whereas 6 descriptors for the QSAR model of SOD activity were RDF120m, Mor27u, R7u+, BELp5, G3m and BELm8. The definition and type of descriptors are listed in Table 2(Tab. 2). The intercorrelation matrix was performed by Pearson's correlation coefficient (r) in which each descriptor was explored using independent descriptors with value of |r| < 0.8 in both RSA and SOD activities (Tables S1 and S2, Supplementary informationexcli2021-4590_supplementary_information.pdf).
The constructed QSAR model for RSA was generated from Eq. (3) using 6 significant descriptors as independent variables, and %RSA as dependent variable. The 13 active sulfonamides (1, 2, 4, 6-15) were used as data set for training and LOO-CV sets to build up and evaluate the QSAR model (Eq. (4).

Table 3(Tab. 3) showed correlation coefficients (R2 and Q2) and root mean square error (RMSE) of training (Tr) set as 0.9948 and 0.0985, respectively, and of leave-one-out cross validation (LOO-CV) set as 0.9708 and 0.5705, respectively. The correlation coefficient in training and LOO-CV sets exhibited high correlation. It indicated reliability of statistical quality with correlation coefficient of training set (R2) > 0.6 and LOO-CV set (Q2) >0.5 (Nantasenamat et al., 2010[22]), but low RMSE values. This suggested that the six descriptors (Table 2(Tab. 2)) were related with antioxidant activity, and their numerical values are presented in Table 4(Tab. 4). The plot between experimental and predicted (%RSA) activity is shown in Figure 2a(Fig. 2). Considering regression coefficients in Eq. (4), descriptors G2u, RDF120m and RDF045m showed positive values of 102.9163, 0.853 and 0.0883, respectively, while Lop, N-067 and H1v descriptors had negative values of -18.7313, -1.0034 and -5.6199, respectively. This implied that the positive values of regression coefficients provided the positive effect which is responsible for increasing the activity, on the other hand, the negative values of regression coefficients reduce the activity.
Similarly, the QSAR model of SOD activity was constructed using 14 active sulfonamides (1-4, 6, 8-16) (Table 1(Tab. 1)), and 6 significant descriptors. The generated QSAR model was obtained as shown in equation (5):

The statistical results of training set displayed R2Tr and RMSETr values of 0.9714 and 0.2306, whereas Q2LOO-CV and RMSELOO-CV of LOO-CV set were 0.8753 and 1.3571, respectively (Table 3(Tab. 3)). The statistical quality provided high correlation between the experimental and the predicted values, but low RMSE values in the training and LOO-CV sets. The six descriptors in Eq. (5) were correlated with the SOD activity, and the numerical values are shown in Table 5(Tab. 5). The plot of experimental and predicted (%SOD) activity is shown in Figure 2b(Fig. 2). The regression coefficients including RDF120m, More27u, BELp5 and BELm8 descriptors have positive values of 2.0192, 18.7889, 4.5407 and 7.3208, respectively, whereas R7u+ and G3m descriptors showed negative values of -13.5517 and -111.5729, respectively. The positive values of regression coefficients impact the increase activity, but the negative values diminished the activity.
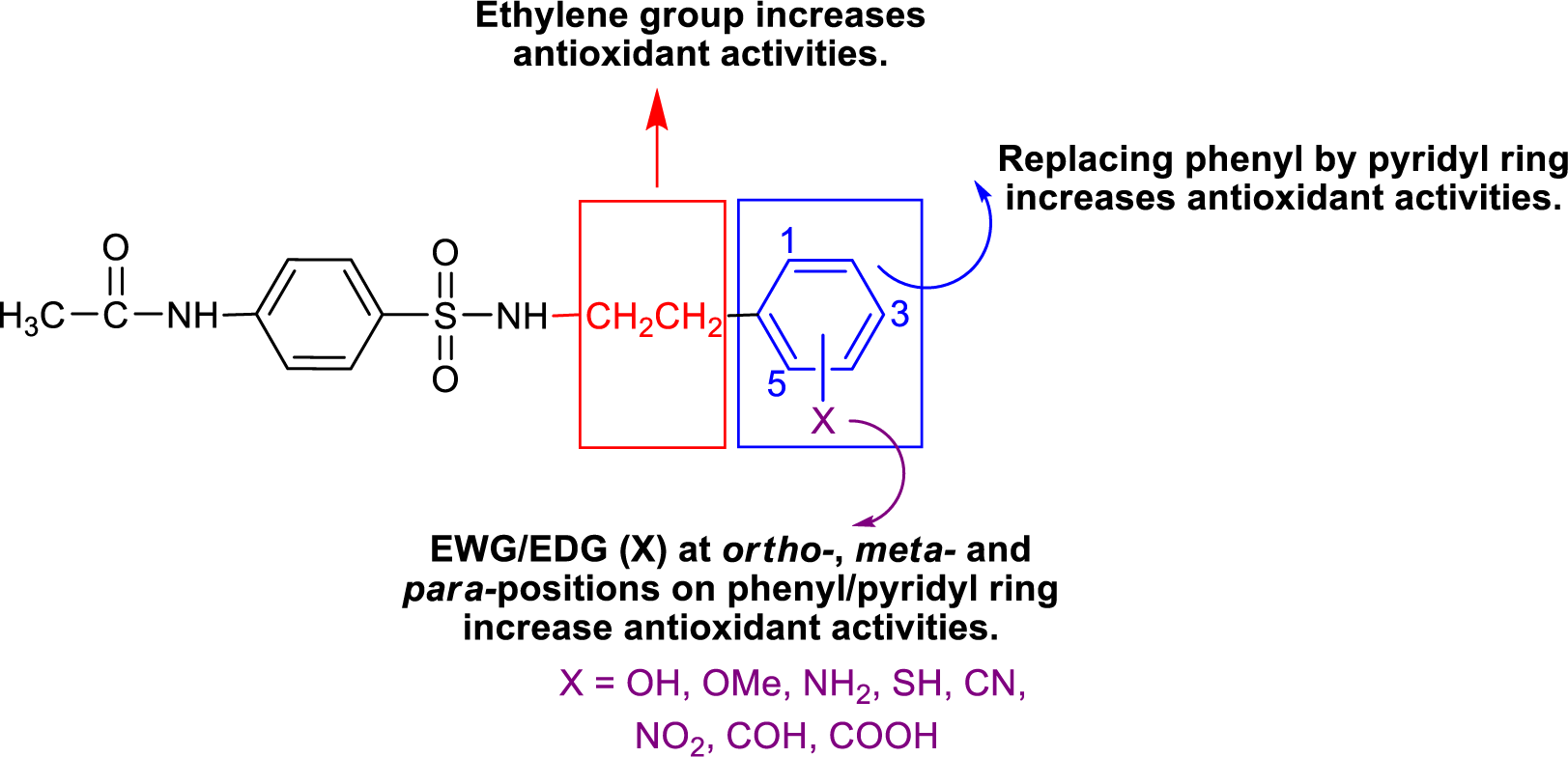
Structure-activity relationship and rational design
Considering the effect of substituted (R) group on antioxidant activity, it was found that R = NHCH2CH2-2-pyridyl (15) exerted the highest RSA (4.62 %) and SOD (38.54 %) activities. When R = N-methyl piperazine, compound 5 displayed no RSA and SOD activities.
Group A compounds (1-4) showed SOD (25.22-30.40 %) and RSA (0.42-1.82 %) activities, compound 2 (R = NHCH2-2-pyridiyl) displayed the highest SOD (30.40 %) but low RSA (1.46 %) activities. Compound 3 (R = NHCH2-3-pyridiyl) as 3-pyridyl analog of compound 2 exerted lower SOD (25.22 %) and no RSA activities. As phenyl (C6H5) analog, compound 1 (R = NHC6H5) showed lower activities of SOD (27.26 %) and RSA (0.42 %) compared with compound 2. Furan compound 4 (R = NHCH2-2-tetrahydrofuranyl) displayed the highest RSA (1.82 %).
The higher SOD activity of 2 could be due to its positive effect of descriptors with higher values of RDF120m (1.472) and Mor27u (-0.054) compared with compound 1 showing lower values of RDF120m (0.000) and Mor27u (-0.070).
For RSA activity, 2 had higher RDF120m (1.472) and G2u (0.177) compared with 1 (RDF120m = 0.000, G2u = 0.175). It should be noted that RDF120m involved in both RSA and SOD activities making 2>1. This high RDF120m value may be resulted from ring N-atom of 2-pyridyl ring of compound 2 compared with phenyl ring of compound 1.
In case of 3-pyridyl ring, 3 (R = NHCH2-3-pyridyl) displayed lower SOD activity with lower RDF120m (0.240) and Mor27u (-0.091) compared with compound 2. This might be due to an isomeric effect of pyridine ring, in which higher atomic polarizabilities (2, BELp5 = 1.167, 3, BELp5 = 1.161) but lower 3rd component symmetry (2, G3m = 0.170, 3, G3m = 0.184) were noted for 2. The R group of ring containing oxygen atom (4) displayed the highest RSA among group A. This could be due to its highest 2nd component symmetry (G2u = 0.187) but the lowest van der Waals volume (H1v = 1.034) (Table 4(Tab. 4)).
Group B compounds 13-16 bearing longer ethylene linker between amino and phenyl/pyridyl rings, compound 15 gave the highest SOD (38.54 %) and RSA (4.62 %) activities, and higher activities than 2-pyridyl analog (2) with shorter CH2 linker.
The QSAR results showed that SOD activity of 15 (R with longer CH2CH2 linker) had the higher values of RDF120m (2.433) and Mor27u (0.082) than 2 containing shorter CH2 linker (RDF120m = 1.472, Mor27u = -0.054). This indicated the effect of higher mass RDF120m of 15 compared with 2. Similar results were noted for RSA (15>2), in which 15 had higher RDF120m = 2.433 and G2u = 0.191 compared with 2 (RDF120m = 1.472, G2u = 0.177).
Similar effect was noted for longer CH2CH2 linker compound 13 (SOD = 30.22 %, RSA = 4.36 %) compared with shorter linker compound 1 (SOD = 27.26 %, RSA = 0.42 %). This could be due to the higher RDF120m (0.205) and Mor27u (-0.006) of 13 in SOD activity compared with 1 (RDF120m = 0.000, Mor27u = -0.070). The similar results of descriptors were seen in RSA of compound 13 (RDF120m = 0.205, G2u = 0.209) and compound 1 (RDF120m = 0.000, G2u = 0.175), in which 13>1.
3,4-Dimethoxyphenyl analog (14) showed higher SOD (32.24 %) activity, but lower RSA (0.82 %) compared with phenyl ring compound 13. The QSAR result (SOD) of 14 showed higher values of RDF120m (2.737), BELp5 (1.393) and BELm8 (1.082) compared with 13 (RDF120m = 0.205, BELp5 = 1.296), BELm8= 1.000). On the other hand, compound 13 with higher RSA displayed high G2u (0.209), low Lop (1.055) whereas low G2u (0.177), high Lop (1.209) were noted for compound 14. High polarizabilities BELp5 of 14 could be due to diOMe groups that enhance the molecule to scavenge superoxide in exerting the SOD activity. High value of G2u symmetry involved in RSA, thus, symmetrical compound 13 (without diOMe) displayed better RSA compared with compound 14.
However, 2-pyridyl compound 15 exerted higher SOD (38.54 %) and RSA (4.62 %) compared with the phenyl analog 13. This might be due to pyridine 15 with N-ring atom displayed SOD activity with higher RDF120m (2.433) compared with 13 (RDF120m = 0.205). Similar results were noted for RSA of 15 (RDF120m = 2.433) compared with 13 (RDF120m = 0.205).
In case of diamino ethane compound 16 (R = NHCH2CH2N(CH2CH3)2), relatively high SOD activity (32.85 %) was noted but the RSA was diminished. Among the investigated compounds, the highest BELp5 (1.422), but the lowest R7u (0.018) were observed for 16. Such high polarizabilities might be due to the effect of polar diamino moieties.
Compounds in group D (10-12) with ring directly linked to the amino group, all displayed RSA and SOD activities. Cyclopentylamino compound (11) showed the highest SOD (31.74 %) but the lowest RSA (0.41 %), whereas cyclohexyl analog (10) exhibited higher RSA (1.29 %) activity. The highest Mor27u (0.158) and the highest BELp5 (1.334) were annotated for the SOD of 11 (R = NHcyclopentyl) compared with 10, (R = NHcyclohexyl, Mor27u = 0.053, BELp5 = 1.236). This could be the ring size effect, as the smaller ring (11) had higher polarizabilities (BELp5) and Mor27u compared with larger ring (10). Compound 10 with higher RSA had high RDF120m (0.148), RDF045m (10.877), but low Lop (1.113) compared with compound 11 having low RDF120m = 0.054, RDF045m = 9.653, but high Lop = 1.144. The higher RSA might be due to high values of mass descriptors of 10 resulting from larger cyclohexyl ring.
Interestingly, in group D compound 12 (R = NHphenyl) had the highest RSA (1.91 %) with the lowest N-067 = 0, but the lowest SOD activity (29.35 %) with the lowest Mor27u (-0.054), BELp5 (0.998) and BELm8 (0.721). This could be due to the ability of phenyl ring that can stabilize the radical species (N•) producing by R group (NH-phenyl) and eventually form resonance stabilized phenyl radical.
Group C compounds (5-9) containing N-heterocyclic ring (R), the results showed that tetrahydroisoquinoline (9) exhibited the highest SOD (29.94 %) and RSA (3.70 %) activities. The highest SOD activity of 9 showed the highest Mor27u (-0.076) but the lowest R7u+ (0.020) among the group (6, 8 and 9). Compound 9 with the highest RSA displayed the highest G2u (0.194) but the lowest H1v (1.288) among the group (6-9).
Compound 5 (R = N-methylpiperazine) was shown to be an inactive antioxidant. The improved activity was noted for N-benzylpiperazine 6 with RSA (2.73 %) and SOD (29.58 %). N-CH3 group of compound 5 was converted to NCH2C6H5 giving rise to compound 6 with improved RSA (2.73 %) and SOD (29.58 %) activities. The SOD activity of compound 6 had the highest RDF120m (0.681), BELp5 (1.409), BELm8 (1.082) but the lowest R7u+ (0.17) among the group (6, 8-9). The RSA of 6 showed the lowest Lop (0.958) among the group (6-9).
In addition, morpholine compound 8 showed both RSA (1.11 %) and SOD (25.80 %) activities. Piperazine bearing bis-sulfonylphenylacetamido moiety (7) exhibited RSA (2.17 %), but no SOD activity. Compound 8 showed the lowest value of G3m (0.163) in SOD activity and the lowest Hlv value (1.135) in RSA among the group (6-9). Compound 7 displayed only the RSA with the highest RDF120m (0.875) and RDF045m (15.337)
To improve the antioxidant activities, the strongest parent antioxidant (15) was rationally designed by modifying its 2-pyridyl moiety as 3- and 4-pyridyl rings. These pyridyls were substituted by electron donating (EDG)/withdrawing (EWG) groups (i.e., OH, OCH3, NH2, SH, CN, NO2, COH and COOH) at various positions on the rings to give a series of new sulfonamides with predicted activities using the constructed QSAR equations (Eqs. 4-5) (Table S3, Supplementary informationexcli2021-4590_supplementary_information.pdf). The obtained values of descriptors for RSA and SOD activities are summarized (Tables S4 and S5, Supplementary informationexcli2021-4590_supplementary_information.pdf).
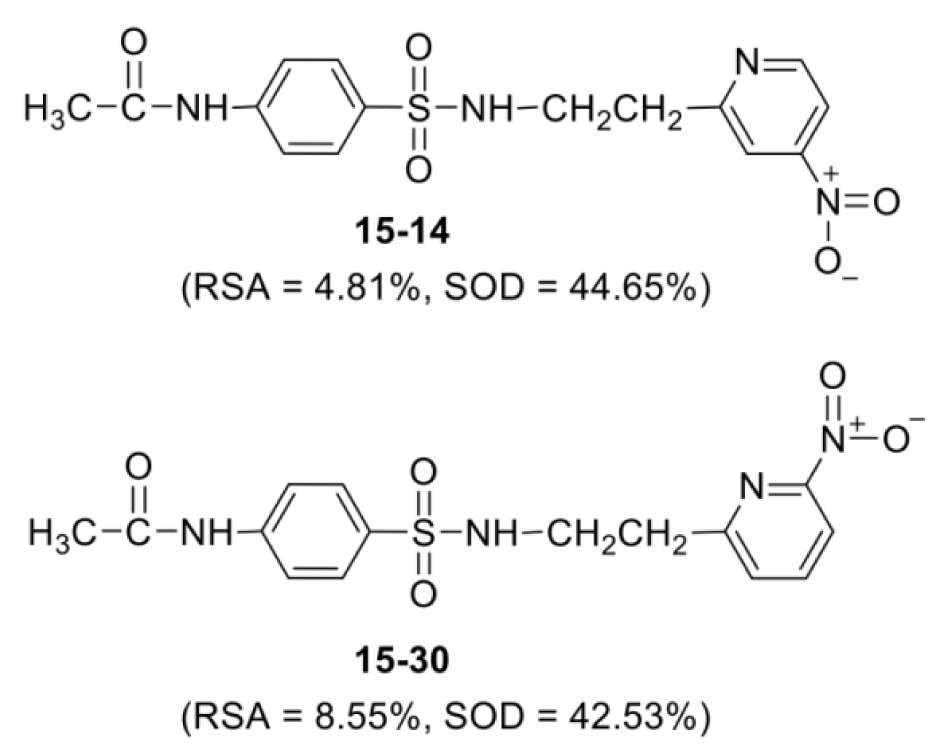
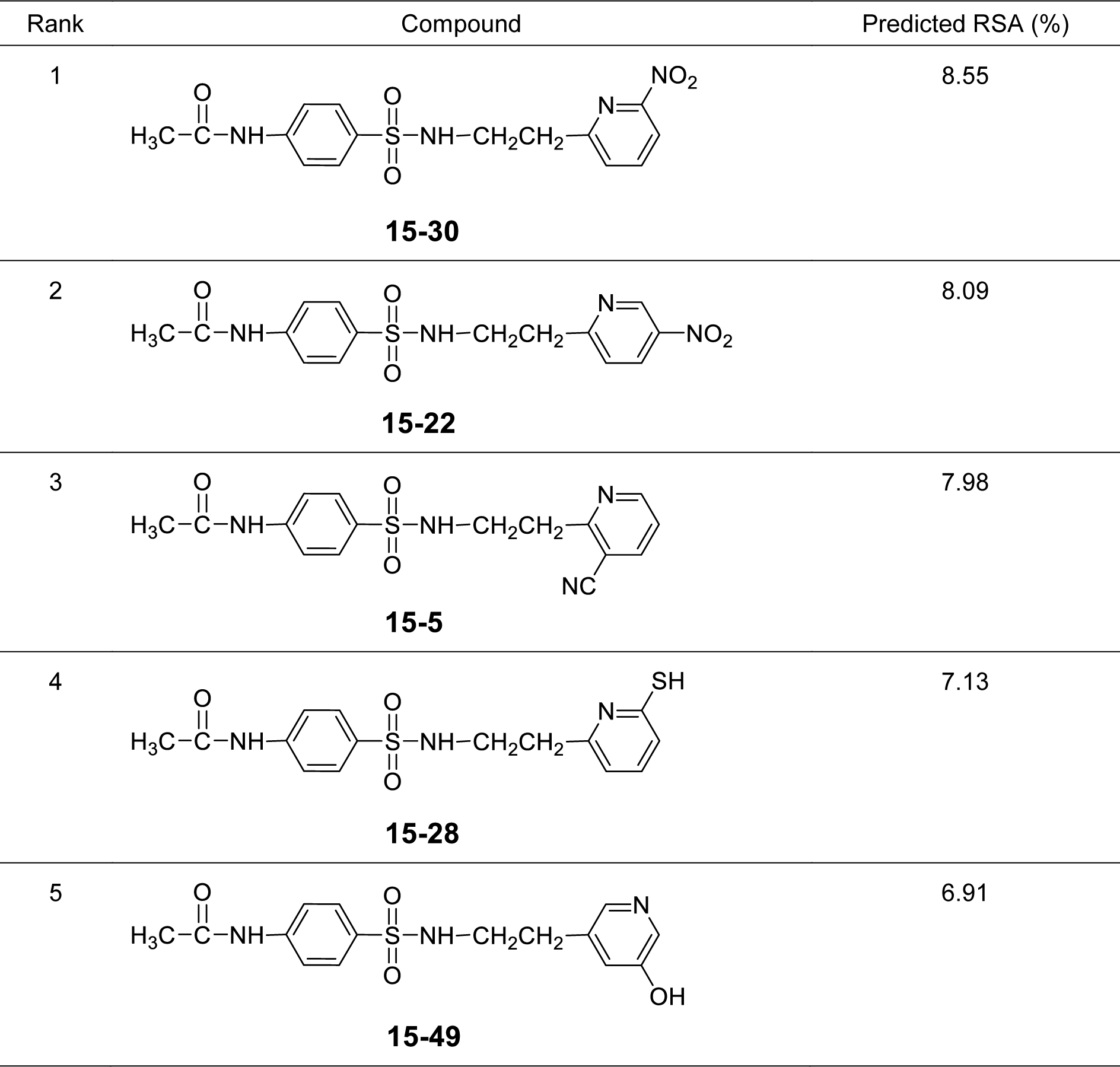
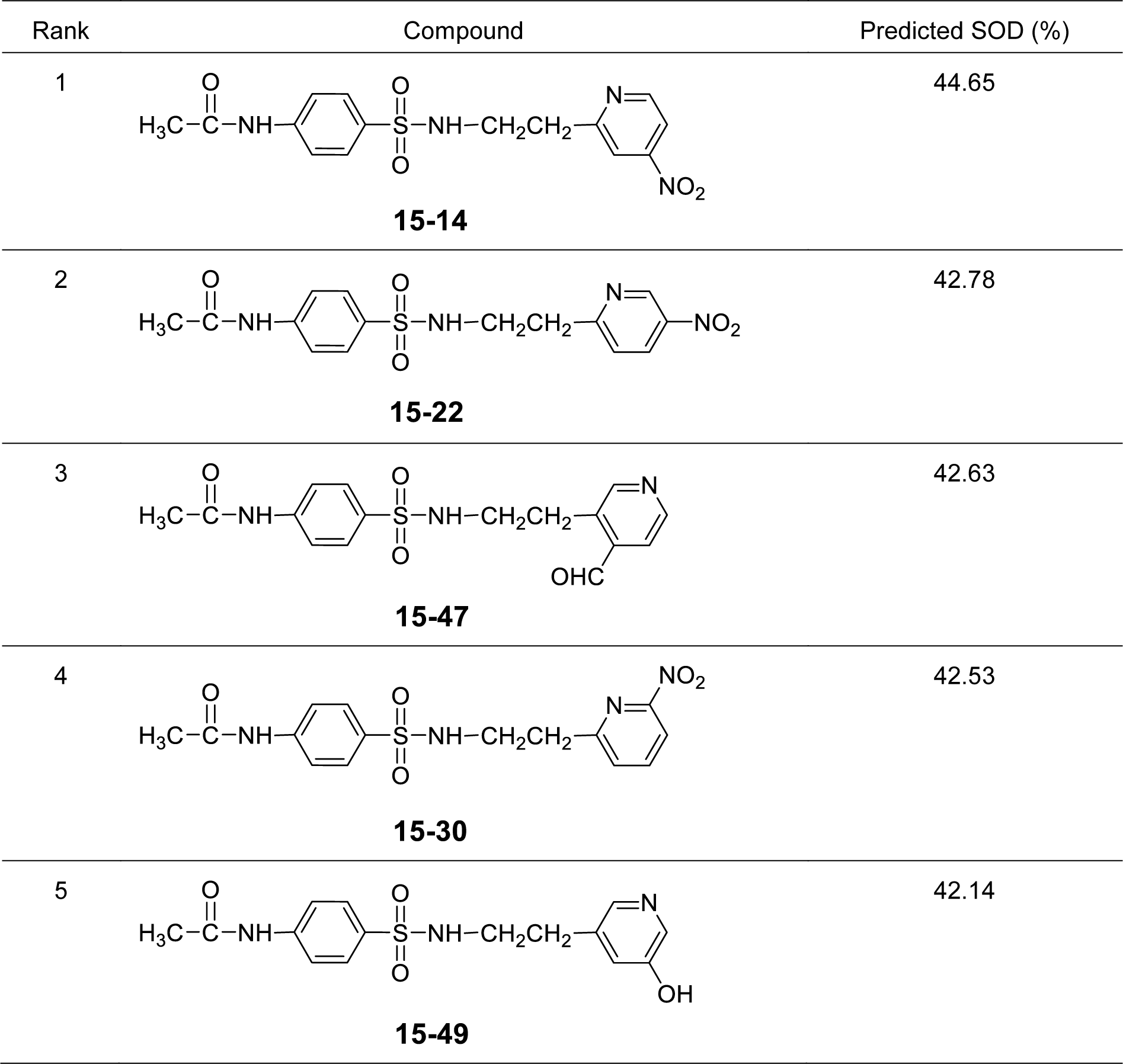
It was found that most of the newly designed sulfonamide derivatives displayed the improved RSA and SOD activities (Table S3, Supplementary informationexcli2021-4590_supplementary_information.pdf) with better molecular descriptor values than the parent compound 15 (Tables 4(Tab. 4) and 5(Tab. 5)). Top five antioxidant sulfonamides were selected from the 80 rationally designed compounds (Table S3, Supplementary informationexcli2021-4590_supplementary_information.pdf) including compounds 15-30 (o-NO2) > 15-22 (m-NO2) > 15-5 (m-CN) > 15-28 (o-SH) > 15-49 (m-OH) with RSA as shown in Table 6(Tab. 6), and compounds with SOD activity (Table 7(Tab. 7)); 15-14 (p-NO2) > 15-22 (m-NO2) > 15-47 (p-COH) > 15-30 (o-NO2) > 15-49 (m-OH).
Most of the active compounds are 2-pyridyl containing electron withdrawing groups (i.e., NO2, CN and CHO) at ortho/para position on ring N-atom. The most active RSA 15-30 (o-NO2) had high values of G2u (0.227), RDF120m (4.311) and RDF045m (11.257), but low H1v (1.138) compared with the parent compound 15 (G2u = 0.191, RDF120m = 2.433, RDF045m = 8.415, H1v = 1.222). As p-NO2 analog (15-14), it displayed the highest SOD activity with high Mor27u (0.190), BELm8 (0.993), BELp5 (1.475), RDF120m (3.787) but low R7u (0.023), and G3m (0.157) compared with the compound 15 (Mor27u = 0.082, BELm8 = 0.926, BELp5 = 1.296, RDF120m = 2.433, R7u = 0.026, G3m = 0.159). The highest SOD activity of 15-14 might be due to high polarizabilities (BELp5 = 1.475) resulting from ionic character of NO2 group (Figure 3(Fig. 3)) and its p-effect on 2-pyridyl ring, which could facilitate superoxide scavenging activity. In addition, o-NO2 (15-30) and m-NO2 (15-22) analogs are also the top five SOD activity (Table 7(Tab. 7)). Particularly, both improved RSA and SOD activities are seen for new designed compounds 15-22 and 15-30. The structure-activity relationship analysis is summarized in Figure 4(Fig. 4).
Drug likeness properties
The investigated sulfonamides were determined for drug likeness characters based on the Lipinski rule of five with the molecular features composed of (1) MW < 500 Da, (2) LogP < 5, (3) nHDon < 5, and (4) nHAcc < 10 using Dragon software. The description of descriptors are as followed: molecular weight (MW) generally represents the molecular size of the compounds, LogP is a parameter to determine molecular hydrophobicity or lipophilicity character of the 1-octanol/water partition coefficient, nHDon and nHAcc were the number of hydrogen bond donors (OH and NH groups) and acceptors (N and O), respectively as well as hydrogen bonding capacity in the molecules (Nantasenamat et al., 2013[23]). It was found that all sulfonamides exhibited drug likeness properties (Table S6, Supplementary informationexcli2021-4590_supplementary_information.pdf). In addition, such drug likeness properties were also noted for the rational design of 80 new sulfonamides (Table S7, Supplementary informationexcli2021-4590_supplementary_information.pdf).
Conclusion
Acetamidosulfonamide derivatives were synthesized and evaluated for their antioxidant and antimicrobial activities. Most sulfonamides exhibited antioxidant activities, particularly, compound 15 exhibited the highest RSA and SOD activities, but all sulfonamides revealed to be inactive antimicrobials. The SAR indicated that the compound bearing 2C chain length with terminal 2-pyridyl ring (15) displayed higher antioxidant activities compared with the corresponding compound with 1C chain (2). Furthermore, the constructed QSAR models displayed important descriptors related with the antioxidant activities (i.e., G2u, Lop, RDF120m, N-067, H1v and RDF045m for RSA, while RDF120m, Mor27u, R7u+, BELp5, G3m and BELm8 for SOD activity. This result led to in silico rational design of 80 new sulfonamide derivatives with electron donating and electron withdrawing groups together with the calculated predicting antioxidant activities using the prototype compound 15. The top five newly designed sulfonamides i.e., nitro compound 15-30 showed the improved antioxidant RSA and SOD activities. In addition, these sulfonamides possessed drug likeness properties. This finding revealed the benefit of constructed QSAR models to insight into the rational design of new antioxidants to be further developed for medicinal applications.
Notes
Apilak Worachartcheewan and Supaluk Prachayasittikul (Center of Data Mining and Biomedical Informatics, Faculty of Medical Technology, Mahidol University, Bangkok 10700, Thailand; E-mail: supaluk@g.swu.ac.th) contributed equally as corresponding author.
Declaration
Supplementary information
Supplementary informationexcli2021-4590_supplementary_information.pdf is available on the EXCLI Journal website.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
AW gratefully acknowledges the research grant supported by the Thailand Research Fund (Grant No. MRG6180053) and the annual budget grant from Mahidol University (B.E. 2562-2563).
References
File-Attachments
- excli2021-4590_supplementary_information.pdf (544,57 KB)
Supplementary information
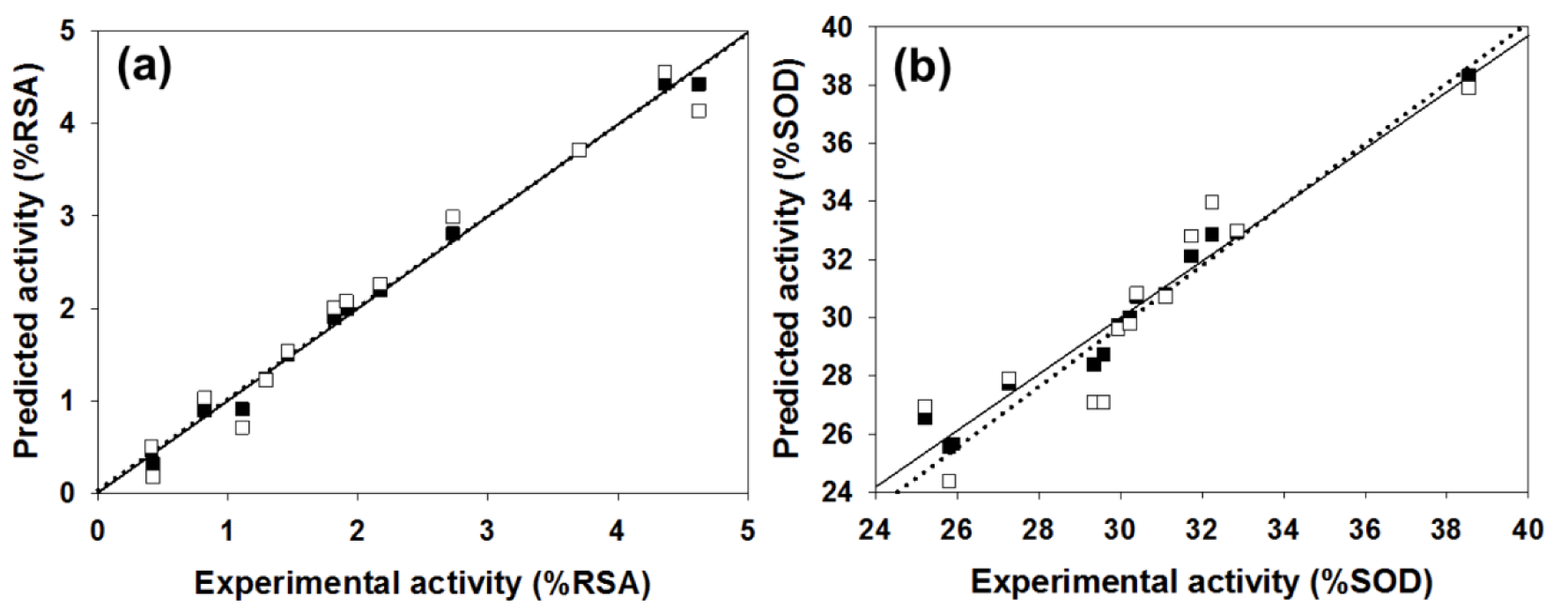
Figure 2: Plot of the experimental versus predicted RSA (a) and SOD (b) activities using MLR method. The training set was represented by black squares and solid line, and the leave-one-out cross-validation set was assigned by white squares and dotted line
[*] Corresponding Author:
Assistant Professor Dr. Apilak Worachartcheewan, Department of Community Medical Technology, Faculty of Medical Technology, Mahidol University, Bangkok 10700, Thailand; Phone: (662) 441-4376, Fax: (662) 441-4380, eMail: apilak.woa@mahidol.ac.th