Machine learning approaches to study the structure-activity relationships of LpxC inhibitors
DOI:
https://doi.org/10.17179/excli2023-6356Keywords:
antimicrobial resistance, LpxC, QSAR, machine learning, cheminformatics, activity cliff, chemotypeAbstract
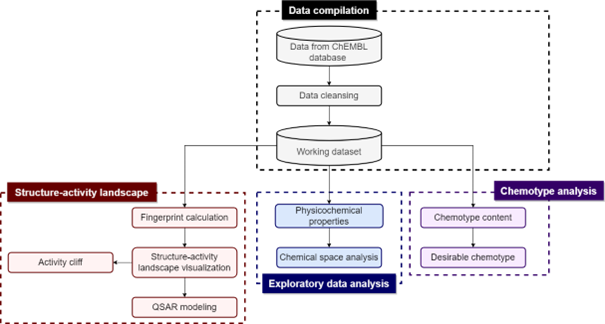
Antimicrobial resistance (AMR) has emerged as one of the global threats to human health in the 21st century. Drug discovery of inhibitors against novel targets rather than conventional bacterial targets has been considered an inevitable strategy for the growing threat of AMR infections. In this study, we applied quantitative structure-activity relationship (QSAR) modeling to the LpxC inhibitors to predict the inhibitory activity. In addition, we performed various cheminformatics analysis consisting of the exploration of the chemical space, identification of chemotypes, performing structure-activity landscape and activity cliffs as well as construction of the Structure-Activity Similarity (SAS) map. We built a total of 24 QSAR classification models using PubChem and MACCS fingerprint with 12 various machine learning algorithms. The best model with PubChem fingerprint is the Extremely Gradient Boost model (accuracy on the training set: 0.937; accuracy on the 10-fold cross-validation set: 0.795; accuracy on the test set: 0.799). Furthermore, it was found that the best model using the MACCS fingerprint was the Random Forest model (accuracy on the training set: 0.955; accuracy on the 10-fold cross-validation set: 0.803; accuracy on the test set: 0.785). In addition, we have identified eight consensus activity cliff generators that are highly informative for further SAR investigations. It is hoped that findings presented herein can provide guidance for further lead optimization of LpxC inhibitors.
Downloads
Additional Files
Published
How to Cite
License
Copyright (c) 2023 Tianshi Yu, Li Chuin Chong, Chanin Nantasenamat, Nuttapat Anuwongcharoen, Theeraphon Piacham

This work is licensed under a Creative Commons Attribution 4.0 International License.
Authors who publish in this journal agree to the following terms:
- The authors keep the copyright and grant the journal the right of first publication under the terms of the Creative Commons Attribution license, CC BY 4.0. This licencse permits unrestricted use, distribution and reproduction in any medium, provided that the original work is properly cited.
- The use of general descriptive names, trade names, trademarks, and so forth in this publication, even if not specifically identified, does not imply that these names are not protected by the relevant laws and regulations.
- Because the advice and information in this journal are believed to be true and accurate at the time of publication, neither the authors, the editors, nor the publisher accept any legal responsibility for any errors or omissions presented in the publication. The publisher makes no guarantee, express or implied, with respect to the material contained herein.
- The authors can enter into additional contracts for the non-exclusive distribution of the journal's published version by citing the initial publication in this journal (e.g. publishing in an institutional repository or in a book).





